A spatially-aware unsupervised pipeline to identify co-methylation regions in DNA methylation data
bioRxiv preprint (posted November 14, 2025)
bioRxiv, New Results, 2025
Abstract
DNA methylation data are high-dimensional and spatially structured, which creates substantial multiple-testing burden and can reduce reproducibility in association analyses.
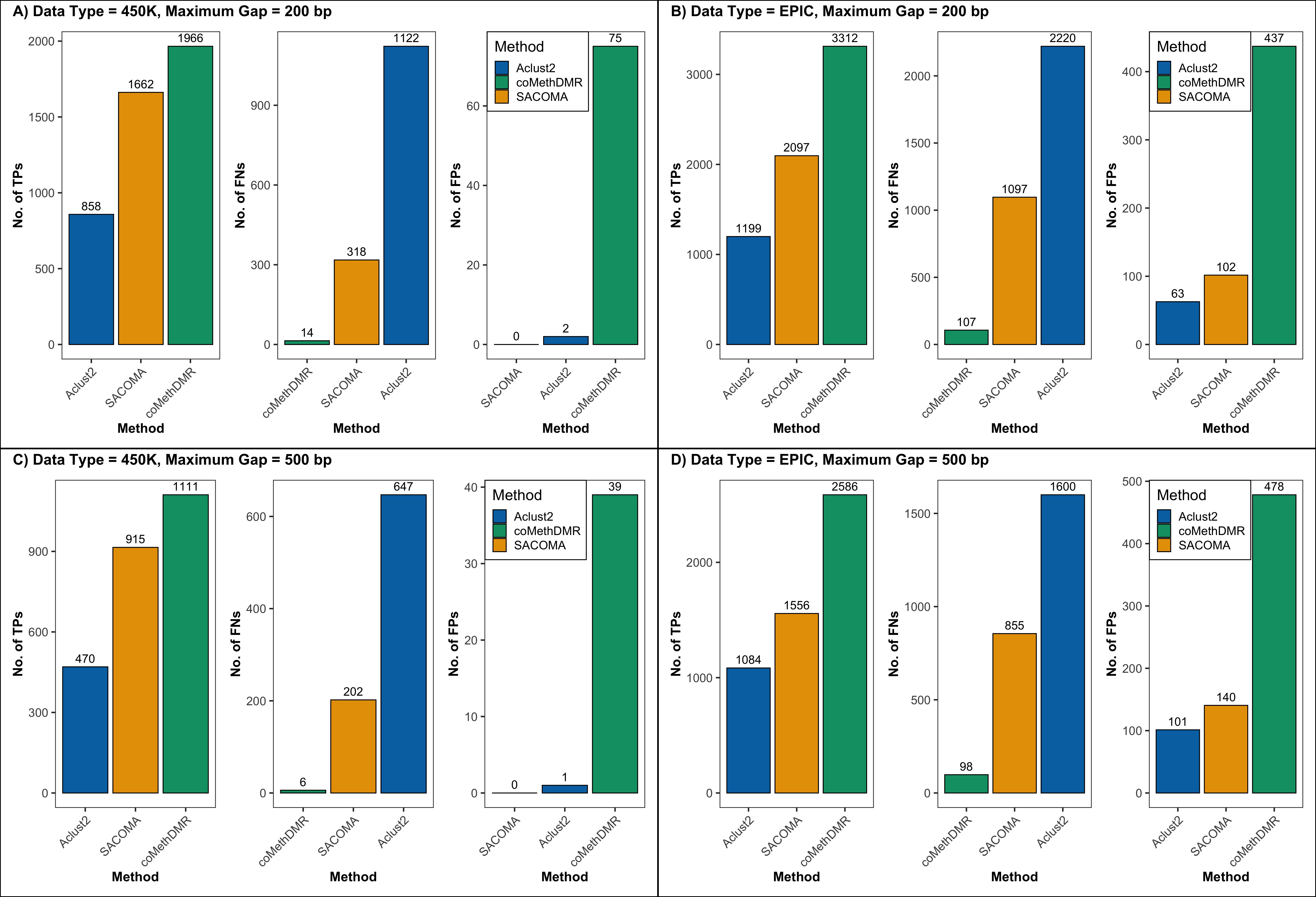
This work introduces SACOMA (Spatially-Aware Clustering for Co-Methylation Analysis), a flexible and unsupervised pipeline that identifies co-methylated regions by combining genomic proximity and methylation similarity through spatially constrained hierarchical clustering.
A tunable, data-adaptive mixing parameter allows SACOMA to avoid rigid threshold assumptions while remaining robust across settings. Although developed for DNAm array studies, the method is generalizable to other spatially dependent data domains.
In simulations and population-level DNAm analyses, SACOMA showed strong sensitivity with effective false-positive control and identified biologically meaningful co-regulated regions, improving both specificity and discovery.
Key Contributions
Spatially-Aware Unsupervised Pipeline
Proposes SACOMA for automatic co-methylation region detection without requiring predefined fixed correlation or distance cutoffs.
Data-Adaptive Mixing
Introduces a tunable mixing parameter to balance spatial proximity and methylation similarity, improving adaptability across datasets.
Improved Statistical Reliability
Demonstrates superior sensitivity with effective false-positive control in extensive simulation experiments compared with existing approaches.
Broader Applicability
Establishes a framework that extends beyond DNAm analyses to any setting involving spatially correlated features.
Citation
BibTeX
@article{meshram2025sacoma,
title={A spatially-aware unsupervised pipeline to identify co-methylation regions in DNA methylation data},
author={Meshram, Siddhant and Fadikar, Arindam and Arunkumar, Ganesan and Chatterjee, Suvo},
journal={bioRxiv},
year={2025},
doi={10.1101/2025.11.13.688356},
note={Preprint}
}Related Publications
- Adaptive Grid-Based Thompson Sampling for Efficient Trajectory Discovery - Complementary method development in model calibration
- bioRxiv record - Latest abstract and version history