Research
My research focuses on developing statistical and computational methods for complex scientific problems, with emphasis on uncertainty quantification, calibration, and scalable inference.
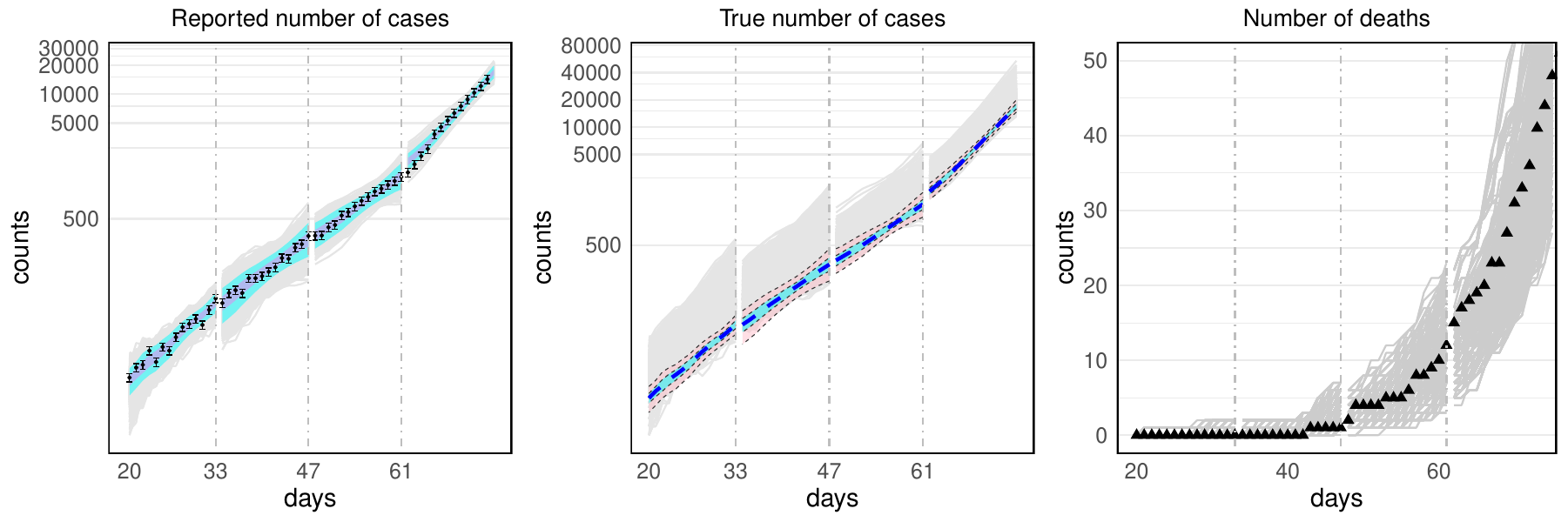
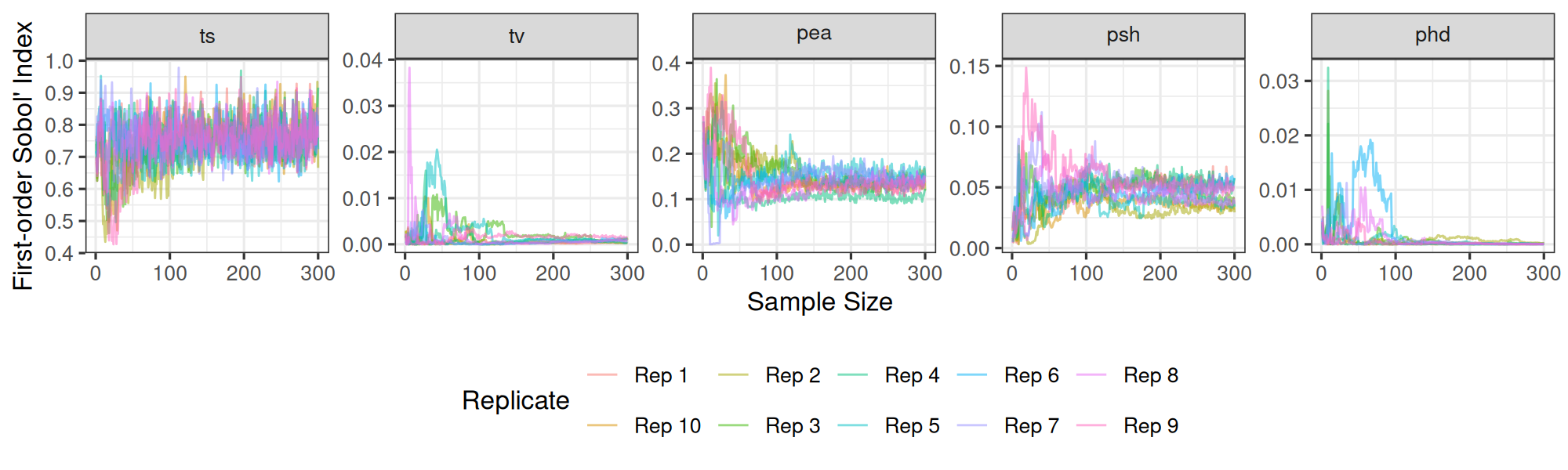
Calibration and UQ
Developing scalable methods for calibrating complex epidemiologic models and quantifying prediction uncertainty. This work combines Gaussian processes, Bayesian inference, and high-performance computing to enable efficient exploration of parameter spaces.
Related Publications
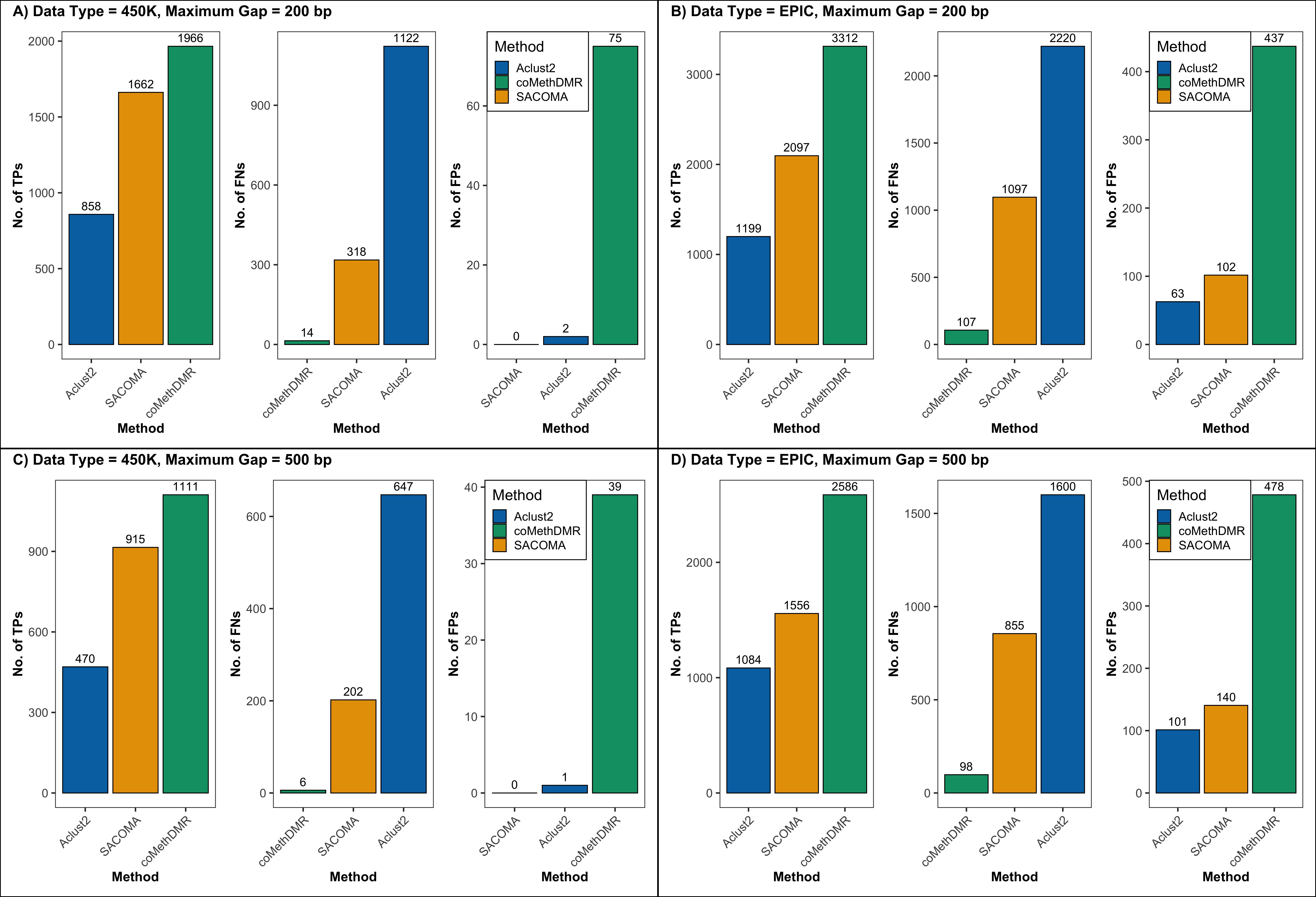
Biostatistics
Statistical methods for high-dimensional omics data, including region-based DNA methylation analysis and robust differential expression inference under heteroscedasticity. This work emphasizes reproducibility, error control, and biologically meaningful discovery.
Related Publications
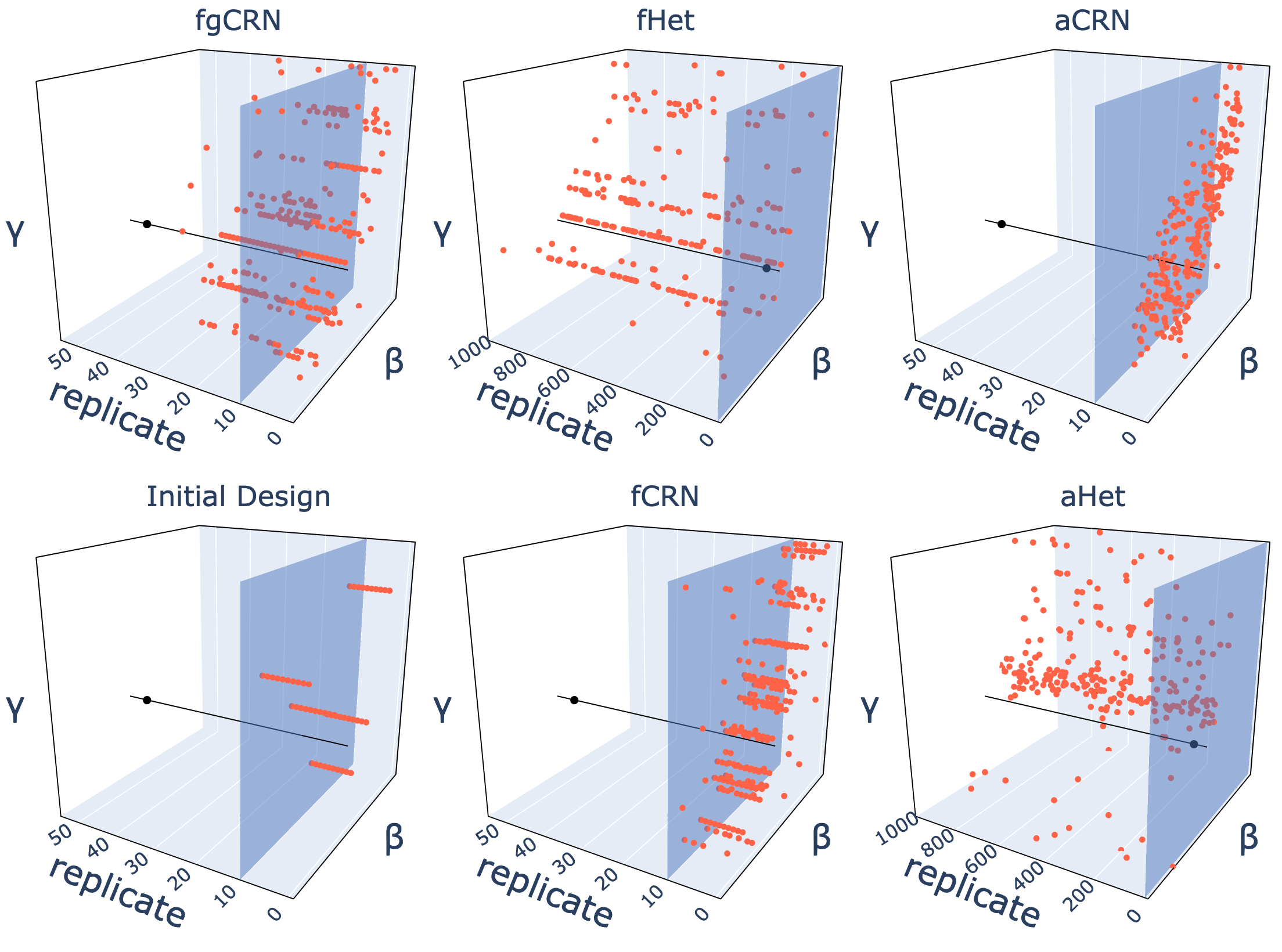
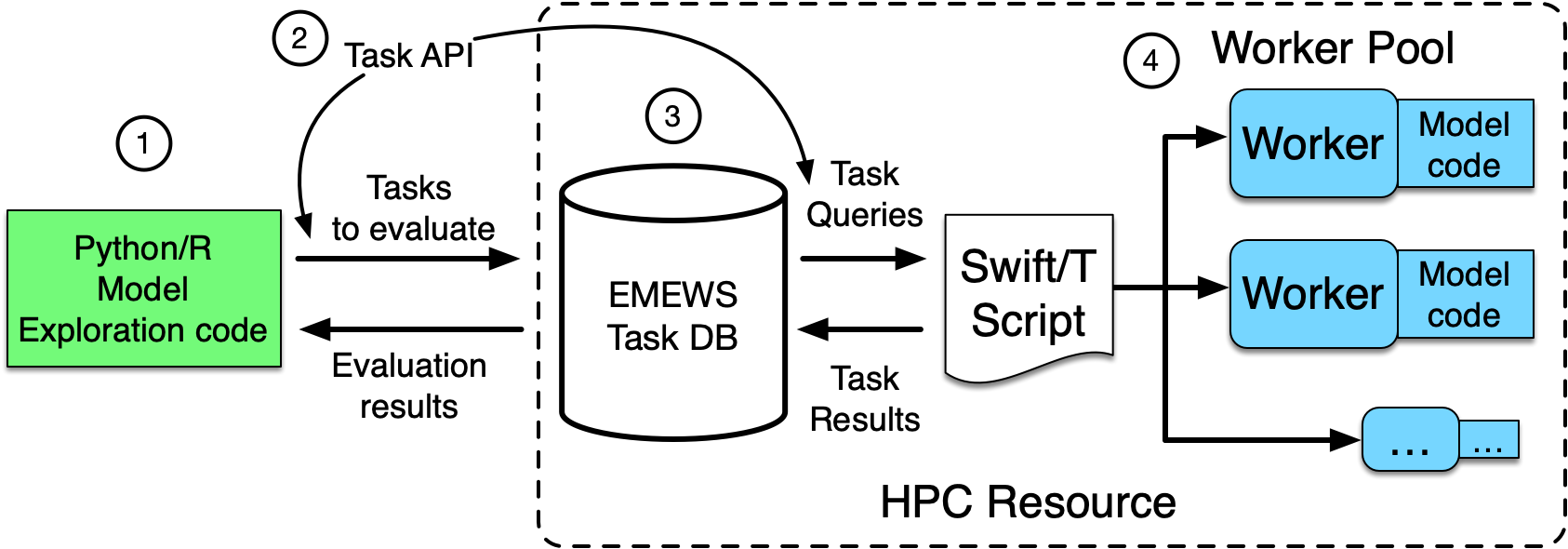
Workflows
Building scalable workflow infrastructures for distributed model exploration, robust epidemic analysis, and reproducible scientific computing on heterogeneous high-performance platforms.